
SUMAC 2.0
8/2016: SUMAC version 2.0 is significantly faster than previous versions due to the use of a new clustering algorithm.
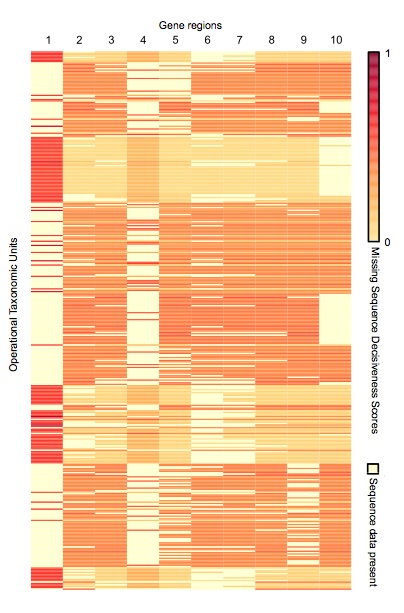
Supermatrix Constructor (SUMAC) is a tool to data-mine GenBank, construct phylogenetic supermatrices, and assess the decisiveness of a matrix given the pattern of missing sequence data. SUMAC calculates a novel metric, Missing Sequence Decisiveness Scores (MSDS), which measure how much each individual missing sequence contributes to the decisiveness of the matrix. MSDS can be used to compare supermatrices and prioritize the acquisition of new sequence data.
SUMAC constructs supermatrices either through an exploratory clustering of all GenBank sequences within a taxonomic group, or by using guide sequences to build homologous clusters in a more targeted manner. SUMAC will assemble supermatrices for any taxonomic group recognized in GenBank, and is optimized to run on multicore processors by utilizing multiple parallel processes. SUMAC is implemented as a Python package that can run as a stand-alone command line program, or its modules and objects can be incorporated within other programs.
SUMAC works on Linux/OSX (not MS Windows), and is available at https://github.com/wf8/sumac under the open source GPLv3 license.
Citation:
Freyman, W.A. 2015. SUMAC: constructing phylogenetic supermatrices and assessing partially decisive taxon coverage. Evolutionary Bioinformatics 2015:11 263-266
[html]
[pdf]